核糖體移碼
核糖體移碼(Ribosomal frameshift)又稱轉譯移碼(Translational frameshift),是生物細胞中核糖體進行轉譯時,mRNA上的特定序列與二級結構使核糖體發生位移而破壞開放閱讀框的現象[3]。由於mRNA上的密碼子是由3個核苷酸對應一個氨基酸,+1、-1等核糖體移碼會影響下游的開放閱讀框,進而轉譯出完全不同的蛋白質[4]。核糖體移碼可使一個mRNA得以轉譯出數種不同的蛋白質產物,此機制主要在病毒(反轉錄病毒[5]、勞斯肉瘤病毒(RSV)[1]、冠狀病毒[6]、流感病毒[7]、反轉錄轉座子等)的mRNA中發現,但也見於一些真核生物細胞基因的mRNA,可能為細胞調控基因表現的機制之一[8][9]。
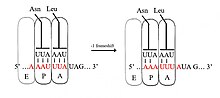

最常見的核糖體移碼為-1核糖體移碼(programmed −1 ribosomal frameshifting, −1 PRF),此外還有較罕見的+1核糖體移碼與-2核糖體移碼[3]。發生-1核糖體移碼的序列通常包含滑動序列、間隔序列(spacer)與莖環等三個元件,典型的滑動序列基序為X_XXY_YYH(X可為任意核苷酸、Y為A或U、H則為ACU三者之一),-1移碼發生時,原與XXY密碼子結合的核糖體P位點與其上的tRNA向前位移,改與XXX結合,同時原與YYH結合的A位點與其tRNA也改與YYY結合,新的反密碼子/密碼子配對除密碼子三號位的核苷酸外,一號位與二號位的核苷酸皆與原本的相同[10],而三號位因有搖擺鹼基對,反密碼子/密碼子的結合力本就較弱,不對兩者的結合造成嚴重影響[3][11]。
發生+1核糖體移碼的序列則沒有特定基序[2],一般機制為使用一較罕見(對應tRNA的量較少)的密碼子使轉譯發生停滯,增加核糖體發生移碼的機會[2][12]。
參考文獻 编辑
- ^ 1.0 1.1 Jacks T, Madhani HD, Masiarz FR, Varmus HE. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. November 1988, 55 (3): 447–458. PMC 7133365 . PMID 2846182. doi:10.1016/0092-8674(88)90031-1.
- ^ 2.0 2.1 2.2 Harger JW, Meskauskas A, Dinman JD. An "integrated model" of programmed ribosomal frameshifting. Trends in Biochemical Sciences. 2002, 27 (9): 448–454. PMID 12217519. doi:10.1016/S0968-0004(02)02149-7.
- ^ 3.0 3.1 3.2 Napthine S, Ling R, Finch LK, Jones JD, Bell S, Brierley I, Firth AE. Protein-directed ribosomal frameshifting temporally regulates gene expression. Nature Communications. 2017, 8: 15582. Bibcode:2017NatCo...815582N. PMC 5472766 . PMID 28593994. doi:10.1038/ncomms15582.
- ^ Atkins JF, Loughran G, Bhatt PR, Firth AE, Baranov PV. Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use. Nucleic Acids Research. 2016, 44 (15): 7007–7078. PMC 5009743 . PMID 27436286. doi:10.1093/nar/gkw530.
- ^ Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988, 331 (6153): 280–283. Bibcode:1988Natur.331..280J. PMID 2447506. S2CID 4242582. doi:10.1038/331280a0.
- ^ Baranov PV, Henderson CM, Anderson CB, Gesteland RF, Atkins JF, Howard MT. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology. 2005, 332 (2): 498–510. PMC 7111862 . PMID 15680415. doi:10.1016/j.virol.2004.11.038 .
- ^ Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science. 2012, 337 (6091): 199–204. Bibcode:2012Sci...337..199J. PMC 3552242 . PMID 22745253. doi:10.1126/science.1222213.
- ^ Ketteler R. On programmed ribosomal frameshifting: the alternative proteomes. Frontiers in Genetics. 2012, 3: 242. PMC 3500957 . PMID 23181069. doi:10.3389/fgene.2012.00242 (英语).
- ^ Advani VM, Dinman JD. Reprogramming the genetic code: The emerging role of ribosomal frameshifting in regulating cellular gene expression. BioEssays. 2016, 38 (1): 21–26. PMC 4749135 . PMID 26661048. doi:10.1002/bies.201500131.
- ^ Brierley I. Ribosomal frameshifting viral RNAs. The Journal of General Virology. 1995,. 76 (Pt 8) (8): 1885–1892. PMID 7636469. doi:10.1099/0022-1317-76-8-1885 .
- ^ Crick FH. Codon—anticodon pairing: the wobble hypothesis. Journal of Molecular Biology. 1966, 19 (2): 548–555. PMID 5969078. doi:10.1016/S0022-2836(66)80022-0.
- ^ Caliskan N, Katunin VI, Belardinelli R, Peske F, Rodnina MV. Programmed −1 frameshifting by kinetic partitioning during impeded translocation. Cell. 2014, 157 (7): 1619–1631. PMC 7112342 . PMID 24949973. doi:10.1016/j.cell.2014.04.041 .