組蛋白乙醯化
組蛋白乙醯化(Histone acetylation)為真核生物染色體上包裹DNA的組蛋白N端之離胺酸被乙醯化的轉譯後修飾,為細胞調控基因表現的一種機制。此反應一般由組蛋白乙醯轉移酶(HAT)催化,组蛋白脱乙酰酶(HDAC)則可催化其逆反應組蛋白脫乙醯化( Histone deacetylation),將組蛋白上的乙醯基水解移除[1]。組蛋白乙醯化是為一種表觀遺傳修飾,乙醯基化會消除組蛋白離胺酸所帶的正電荷,使其與DNA(帶負電)的結合力降低[2],因此可將原本纏繞較緊密的染色體結構(異染色質)轉成較疏鬆的型態(真染色質),有利於轉錄的進行而提升基因表現;組蛋白脫乙醯化的功能則與之相反,可使染色體結構變得更緊密而降低基因表現[3][4]。
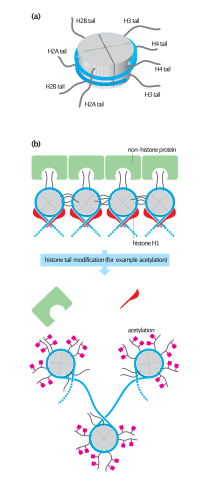
(a)核小體示意圖,可見組蛋白的N端向外延伸
(b)組蛋白未被乙醯化的狀態,其N端與周圍的組蛋白或其他蛋白(綠色結合),染色體包裹較緊密
(c)組蛋白被乙醯化的狀態,染色體包裹較鬆散,有利於基因表現
組蛋白上不同位點的多種修飾(包括乙醯化、甲基化、磷酸化等)可能組合成組蛋白密碼,共同影響染色體結構,並與細胞中的其他蛋白結合以調控基因的轉錄[5][6],例如具有布罗莫结构域的蛋白(包括許多轉錄因子)可與被乙醯化的組蛋白結合[7] 。組蛋白密碼與DNA甲基化皆為表觀遺傳密碼的一部份[8]。
組蛋白乙醯化調控許多基因的表現,因此其調控異常與發炎性疾病、心血管疾病、神經性疾病和多種癌症相關[9][10]。此外對菸酒和藥物的成癮機制可能也與其促進染色體特定區域的組蛋白乙醯化以影響基因表現有關[11][12]。
參考文獻 编辑
- ^ Grunstein M. Histone acetylation in chromatin structure and transcription (PDF). Nature. September 1997, 389 (6649): 349–52 [2021-04-24]. Bibcode:1997Natur.389..349G. PMID 9311776. doi:10.1038/38664. (原始内容存档 (PDF)于2016-03-03).
- ^ Watson, James D.; Baker, Tania A.; Gann, Alexander; Levine, Michael; Losik, Richard. Molecular biology of the gene Seventh. Boston: Pearson/CSH Press. 2014. ISBN 978-0-321-76243-6.
- ^ Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes & Development. March 1998, 12 (5): 599–606. PMID 9499396. doi:10.1101/gad.12.5.599.
- ^ de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. The Biochemical Journal. March 2003, 370 (Pt 3): 737–49. PMC 1223209 . PMID 12429021. doi:10.1042/BJ20021321.
- ^ Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nature Reviews. Cancer. July 2010, 10 (7): 457–69. PMC 3262678 . PMID 20574448. doi:10.1038/nrc2876.
- ^ Barratt MJ, Hazzalin CA, Cano E, Mahadevan LC. Mitogen-stimulated phosphorylation of histone H3 is targeted to a small hyperacetylation-sensitive fraction. Proceedings of the National Academy of Sciences of the United States of America. May 1994, 91 (11): 4781–5. Bibcode:1994PNAS...91.4781B. PMC 43872 . PMID 8197135. doi:10.1073/pnas.91.11.4781.
- ^ Sanchez R, Zhou MM. The role of human bromodomains in chromatin biology and gene transcription. Current Opinion in Drug Discovery & Development. September 2009, 12 (5): 659–65. PMC 2921942 . PMID 19736624.
- ^ Jenuwein T, Allis C. Translating the histone code. Science. 2001, 293 (5532): 1074–80. PMID 11498575. doi:10.1126/science.1063127.
- ^ Barnes PJ, Adcock IM, Ito K. Histone acetylation and deacetylation: importance in inflammatory lung diseases. The European Respiratory Journal. March 2005, 25 (3): 552–63. PMID 15738302. doi:10.1183/09031936.05.00117504.
- ^ Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. August 2007, 26 (37): 5420–32. PMID 17694083. doi:10.1038/sj.onc.1210610.
- ^ Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nature Reviews. Neuroscience. October 2011, 12 (11): 623–37. PMC 3272277 . PMID 21989194. doi:10.1038/nrn3111.
- ^ Hitchcock LN, Lattal KM. Histone-mediated epigenetics in addiction. Epigenetics and Neuroplasticity—Evidence and Debate. Progress in Molecular Biology and Translational Science 128. 2014: 51–87. ISBN 9780128009772. PMC 5914502 . PMID 25410541. doi:10.1016/B978-0-12-800977-2.00003-6.
|journal=被忽略 (帮助)