单细胞分析
| 此条目可参照英语维基百科相应条目来扩充。 (2020年1月15日) |
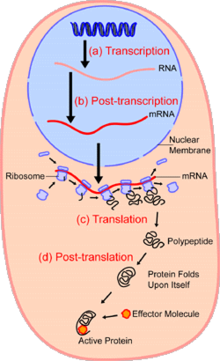
在细胞生物学中,单细胞分析(英语:single-cell analysis)是在单一细胞层级上的基因体学(genomics)、转录体学(transcriptomics)、蛋白质体学(proteomics)与代谢体学(metabolomics)的研究。[1][2]因为原核与真核细胞的族群皆有异质性,所以分析单一细胞可能发现研究整个细胞族群时无法观测的机制。[3] 萤光激发的细胞分选(FACS)之类的科技可将被选取的单一细胞从复杂的样本中精确地分离出来,而高通量的单一细胞分割的科技[4][5]可以同时执行数百或数千个单一未分选的细胞的分子分析,这对分析基因型相同的细胞群中总转录本(transcriptome)的变异特别有用,使人们可以定义以其他方法无法侦测的细胞子类型。新科技的发展使人们更有能力分析单一细胞的基因体(genome)与总转录本,还有量化它的蛋白质体(proteome)与代谢体(metabolome)。[6][7][8]质谱法的一些新发展已经成为用来做单细胞的蛋白质体与代谢体分析的重要分析工具。[9][10]原位定序与萤光原位杂交(FISH)不要求分离细胞,这些技术越来越常被用来做组织的分析。[11]
参考文献 编辑
- ^ Wang D, Bodovitz S. Single cell analysis: the new frontier in 'omics'. Trends in Biotechnology. June 2010, 28 (6): 281–90. PMC 2876223 . PMID 20434785. doi:10.1016/j.tibtech.2010.03.002.
- ^ Habibi I, Cheong R, Lipniacki T, Levchenko A, Emamian ES, Abdi A. Computation and measurement of cell decision making errors using single cell data. PLoS Computational Biology. April 2017, 13 (4): e1005436. PMC 5397092 . PMID 28379950. doi:10.1371/journal.pcbi.1005436.
- ^ Altschuler SJ, Wu LF. Cellular heterogeneity: do differences make a difference?. Cell. May 2010, 141 (4): 559–63 [2020-01-17]. PMC 2918286 . PMID 20478246. doi:10.1016/j.cell.2010.04.033. (原始内容存档于2017-04-19).
- ^ Mora-Castilla, S., et al. "Miniaturization technologies for efficient single-cell library preparation for next-generation sequencing" Journal of Laboratory Automation (2016): 557–67, https://doi.org/10.1177/2211068216630741
- ^ Zheng GX et al, Nat Commun. 2017, “Massively parallel digital transcriptional profiling of single cells”, DOI:10.1038/ncomms14049 (页面存档备份,存于互联网档案馆)
- ^ Huang L, Ma F, Chapman A, Lu S, Xie XS. Single-Cell Whole-Genome Amplification and Sequencing: Methodology and Applications. Annual Review of Genomics and Human Genetics. 2015, 16 (1): 79–102. PMID 26077818. doi:10.1146/annurev-genom-090413-025352.
- ^ Wu AR, Wang J, Streets AM, Huang Y. Single-Cell Transcriptional Analysis. Annual Review of Analytical Chemistry. June 2017, 10 (1): 439–462. PMID 28301747. doi:10.1146/annurev-anchem-061516-045228.
- ^ Tsioris K, Torres AJ, Douce TB, Love JC. A new toolbox for assessing single cells. Annual Review of Chemical and Biomolecular Engineering. 2014, 5: 455–77. PMC 4309009 . PMID 24910919. doi:10.1146/annurev-chembioeng-060713-035958.
- ^ Comi TJ, Do TD, Rubakhin SS, Sweedler JV. Categorizing Cells on the Basis of their Chemical Profiles: Progress in Single-Cell Mass Spectrometry. Journal of the American Chemical Society. March 2017, 139 (11): 3920–3929. PMC 5364434 . PMID 28135079. doi:10.1021/jacs.6b12822.
- ^ Zhang L, Vertes A. Single-Cell Mass Spectrometry Approaches to Explore Cellular Heterogeneity. Angewandte Chemie. April 2018, 57 (17): 4466–4477. PMID 29218763. doi:10.1002/anie.201709719.
- ^ Lee JH. De Novo Gene Expression Reconstruction in Space. Trends in Molecular Medicine. July 2017, 23 (7): 583–593. PMC 5514424 . PMID 28571832. doi:10.1016/j.molmed.2017.05.004.