DNA结合蛋白
DNA结合蛋白(DNA-binding protein)是指能透过DNA结合结构域(DBD)与单链或双链DNA结合的一类蛋白质[1][2][3]。一般来说,能与特异DNA序列结合的DNA结合蛋白主要经由与B-DNA(生物体内DNA一般都是B-DNA形式)的大沟结合来识别DNA序列。因为B-DNA的大沟区域能容许更多蛋白质上的官能团靠近并与之进行相互作用[4][5]。

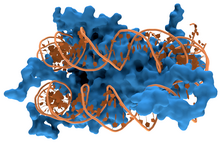
蛋白质与DNA的相互作用
编辑蛋白质与DNA的结合受到pH值、温度、离子强度、电场,以及大分子拥挤等多种条件的影响,具体分为特异性与非特异性两种。非特异性的结合指蛋白质与DNA的结合不依赖特定序列,而特异性的结合则只会发生在特定的DNA序列上[6][7]。在Cas9/CRSIPER系统出现前作为主流基因编辑工具的锌指蛋白以及TALENs都是依赖特异性的DNA蛋白间结合工作的[8]。
检测方法
编辑较早期的检测蛋白与DNA间相互作用的方法[9]包括凝胶迁移实验(EMSA)[10][11]、DNA-蛋白质相互作用ELISA实验(DPI-ELISA)[12]、DNA酶足迹法(DNase footprinting assay) [13]等等。而染色质免疫沉淀法(ChIP)则是目前较为通行的检测体内DNA与蛋白质相互作用的方法[14]。
非特异性DNA结合蛋白
编辑最常见的一种非特异性DNA结合蛋白是真核生物染色质中与DNA结合的组蛋白。组蛋白主要是通过所带的正电荷与带负电的DNA结合,所以组蛋白与DNA之间的结合不依赖DNA上的特定序列。这也符合组蛋白的生物学功能,因为组蛋白需要与各种不同的DNA片段结合形成染色质的基本组成单位核小体[15][16]。另外,真核生物染色质上能够使DNA链发生弯曲的高迁移率族(HMG)蛋白与DNA之间的结合也是非特异性的[17][18][19]。
特异性DNA结合蛋白
编辑很多特异性DNA结合蛋白属于转录因子。转录因子依赖其特有的DNA结合结构域与DNA上的特定序列结合。一般来说,转录因子在与DNA结合后,会招募酶对DNA或组蛋白进行修饰,进而调控基因的表达水平[20][21]。
参见
编辑参考文献
编辑- ^ Travers, A. A. DNA-protein interactions. London: Springer. 1993. ISBN 978-0-412-25990-6.
- ^ Pabo CO, Sauer RT. Protein-DNA recognition. Annu. Rev. Biochem. 1984, 53 (1): 293–321. PMID 6236744. doi:10.1146/annurev.bi.53.070184.001453.
- ^ Dickerson R.E. The DNA helix and how it is read. Sci Am. 1983, 249 (6): 94–111. Bibcode:1983SciAm.249f..94D. doi:10.1038/scientificamerican1283-94.
- ^ Zimmer C, Wähnert U. Nonintercalating DNA-binding ligands: specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material. Prog. Biophys. Mol. Biol. 1986, 47 (1): 31–112. PMID 2422697. doi:10.1016/0079-6107(86)90005-2 .
- ^ Dervan PB. Design of sequence-specific DNA-binding molecules. Science. April 1986, 232 (4749): 464–71. Bibcode:1986Sci...232..464D. PMID 2421408. doi:10.1126/science.2421408.
- ^ Bewley CA, Gronenborn AM, Clore GM. Minor groove-binding architectural proteins: structure, function, and DNA recognition. Annu Rev Biophys Biomol Struct. 1998, 27: 105–31. PMC 4781445 . PMID 9646864. doi:10.1146/annurev.biophys.27.1.105.
- ^ Ganji, Mahipal; Docter, Margreet; Le Grice, Stuart F. J.; Abbondanzieri, Elio A. DNA binding proteins explore multiple local configurations during docking via rapid rebinding. Nucleic Acids Research. 2016-09-30, 44 (17): 8376–8384. ISSN 0305-1048. PMC 5041478 . PMID 27471033. doi:10.1093/nar/gkw666.
- ^ Clark KJ, Voytas DF, Ekker SC. A TALE of two nucleases: gene targeting for the masses?. Zebrafish. September 2011, 8 (3): 147–9. PMC 3174730 . PMID 21929364. doi:10.1089/zeb.2011.9993.
- ^ Cai YH, Huang H. Advances in the study of protein–DNA interaction. Amino Acids. July 2012, 43 (3): 1141–6. PMID 22842750. S2CID 310256. doi:10.1007/s00726-012-1377-9.
- ^ Fried M, Crothers DM. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981, 9 (23): 6505–6525. PMC 327619 . PMID 6275366. doi:10.1093/nar/9.23.6505.
- ^ Garner MM, Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981, 9 (13): 3047–3060. PMC 327330 . PMID 6269071. doi:10.1093/nar/9.13.3047.
- ^ Brand LH, Kirchler T, Hummel S, Chaban C, Wanke D. DPI-ELISA: a fast and versatile method to specify the binding of plant transcription factors to DNA in vitro.. Plant Methods. 2010, 25 (6): 25. PMC 3003642 . PMID 21108821. doi:10.1186/1746-4811-6-25 .
- ^ Galas DJ, Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978, 5 (9): 3157–3170. PMC 342238 . PMID 212715. doi:10.1093/nar/5.9.3157.
- ^ Collas, Philippe. The Current State of Chromatin Immunoprecipitation. Molecular Biotechnology. January 2010, 45 (1): 87–100. PMID 20077036. doi:10.1007/s12033-009-9239-8.
- ^ Luger K, Mäder A, Richmond R, Sargent D, Richmond T. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997, 389 (6648): 251–60. Bibcode:1997Natur.389..251L. PMID 9305837. S2CID 4328827. doi:10.1038/38444.
- ^ Jenuwein T, Allis C. Translating the histone code. Science. 2001, 293 (5532): 1074–80. CiteSeerX 10.1.1.453.900 . PMID 11498575. S2CID 1883924. doi:10.1126/science.1063127.
- ^ Thomas J. HMG1 and 2: architectural DNA-binding proteins. Biochem Soc Trans. 2001, 29 (Pt 4): 395–401. PMID 11497996. doi:10.1042/BST0290395.
- ^ Murugesapillai, Divakaran; McCauley, Micah J.; Huo, Ran; Nelson Holte, Molly H.; Stepanyants, Armen; Maher, L. James; Israeloff, Nathan E.; Williams, Mark C. DNA bridging and looping by HMO1 provides a mechanism for stabilizing nucleosome-free chromatin. Nucleic Acids Research. 2014, 42 (14): 8996–9004. PMC 4132745 . PMID 25063301. doi:10.1093/nar/gku635.
- ^ Murugesapillai, Divakaran; McCauley, Micah J.; Maher, L. James; Williams, Mark C. Single-molecule studies of high-mobility group B architectural DNA bending proteins. Biophysical Reviews. 2017, 9 (1): 17–40. PMC 5331113 . PMID 28303166. doi:10.1007/s12551-016-0236-4.
- ^ Myers L, Kornberg R. Mediator of transcriptional regulation. Annu Rev Biochem. 2000, 69 (1): 729–49. PMID 10966474. doi:10.1146/annurev.biochem.69.1.729.
- ^ Li Z, Van Calcar S, Qu C, Cavenee W, Zhang M, Ren B. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc Natl Acad Sci USA. 2003, 100 (14): 8164–9. Bibcode:2003PNAS..100.8164L. PMC 166200 . PMID 12808131. doi:10.1073/pnas.1332764100 .