核糖体停滞
核糖体停滞(Ribosomal pause, Ribosomal stall)[1]是细胞中核糖体翻译mRNA时发生停滞的现象,在原核生物与真核生物细胞中皆会发生[2][3]。核糖体分析等实验技术可用于找出mRNA中发生核糖体停滞的位点[4]。
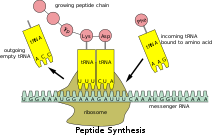
机制 编辑
早在1980年代即有研究显示核糖体翻译mRNA不同区域的的速率不一致,当时认为翻译较慢的位点是因含有罕见的密码子,对应的tRNA在细胞中含量较少,因此与mRNA结合所需的时间较长[5]。但近年核糖体分析的实验结果显示核糖体发生停滞的位点与其对应tRNA的含量不一定有关,意即有些核糖体停滞并非由罕见密码子造成[2]。除此之外造成核糖体停滞的原因还有多脯氨酸序列(polyproline)、tRNA未激活(未连接氨基酸)等[1],此类停滞为可逆,可由eIF5A(真核生物)或EFP(原核生物)蛋白解决,为细胞调控翻译的机制之一,除控制蛋白质翻译的产量[6],还可能与蛋白质折叠有关(即在蛋白质的结构域翻译结束时发生停滞,让其有时间完成折叠)[7],或者促使核糖体移码发生[8]。缺少eIF5A的真核细胞发生核糖体停滞的频率会增加[9]。
有时核糖体停滞为不可逆,原核生物与真核生物皆有将核糖体自mRNA释出的机制。当真核细胞中mRNA上不具终止密码子时,核糖体翻译后会停滞于mRNA的末端,此时细胞会启动无终止密码子媒介式分解途径(NSD)将核糖体释出,并将该mRNA与翻译的多肽产物降解;有时核糖体会遇到mRNA上较复杂的二级结构而停滞,细胞则可启动翻译停滞分解(no-go decay,NGD)途径,亦可将核糖体释出,并降解mRNA及多肽产物[8]。细菌则可以转运-信使RNA(tmRNA)启动反式翻译来处理停滞的核糖体[1],部分细菌还有ArfA、ArfB等其他途径[1][10]。
参考文献 编辑
- ^ 1.0 1.1 1.2 1.3 Buskirk, Allen R.; Green, Rachel. Ribosome pausing, arrest and rescue in bacteria and eukaryotes. Philosophical Transactions of the Royal Society B: Biological Sciences. 2017, 372 (1716): 20160183. PMC 5311927 . PMID 28138069. doi:10.1098/rstb.2016.0183.
- ^ 2.0 2.1 Li GW, Oh E, Weissman JS. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012, 484 (7395): 538–41 [2021-12-06]. Bibcode:2012Natur.484..538L. PMC 3338875 . PMID 22456704. doi:10.1038/nature10965. (原始内容存档于2021-12-06).
- ^ Lopinski JD, Dinman JD, Bruenn JA. Kinetics of ribosomal pausing during programmed -1 translational frameshifting. Molecular and Cellular Biology. 2000, 20 (4): 1095–103. PMC 85227 . PMID 10648594. doi:10.1128/MCB.20.4.1095-1103.2000.
- ^ Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012, 335 (6068): 552–7. Bibcode:2012Sci...335..552B. PMC 3414261 . PMID 22194413. doi:10.1126/science.1215110.
- ^ Kontos H, Napthine S, Brierley I. Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Molecular and Cellular Biology. 2001, 21 (24): 8657–70. PMC 100026 . PMID 11713298. doi:10.1128/MCB.21.24.8657-8670.2001.
- ^ Darnell AM, Subramaniam AR, O'Shea EK. Translational Control through Differential Ribosome Pausing during Amino Acid Limitation in Mammalian Cells. Molecular Cell. 2018, 71 (2): 229–243.e11. PMC 6516488 . PMID 30029003. doi:10.1016/j.molcel.2018.06.041.
- ^ Gawroński P, Jensen PE, Karpiński S, Leister D, Scharff LB. Pausing of Chloroplast Ribosomes Is Induced by Multiple Features and Is Linked to the Assembly of Photosynthetic Complexes. Plant Physiology. March 2018, 176 (3): 2557–2569. PMC 5841727 . PMID 29298822. doi:10.1104/pp.17.01564.
- ^ 8.0 8.1 Buchan JR, Stansfield I. Halting a cellular production line: responses to ribosomal pausing during translation. Biology of the Cell. 2007, 99 (9): 475–87. PMID 17696878. doi:10.1042/BC20070037 .
- ^ Manjunath H, Zhang H, Rehfeld F, Han J, Chang TC, Mendell JT. Suppression of Ribosomal Pausing by eIF5A Is Necessary to Maintain the Fidelity of Start Codon Selection. Cell Reports. 2019, 29 (10): 3134–3146.e6. PMC 6917043 . PMID 31801078. doi:10.1016/j.celrep.2019.10.129.
- ^ Chan, KH; Petrychenko, V; Mueller, C; Maracci, C; Holtkamp, W; Wilson, DN; Fischer, N; Rodnina, MV. Mechanism of ribosome rescue by alternative ribosome-rescue factor B.. Nature Communications. 2020, 11 (1): 4106. PMC 7427801 . PMID 32796827. doi:10.1038/s41467-020-17853-7.